Computational Tool Provides New Understanding into Huntington’s Disease
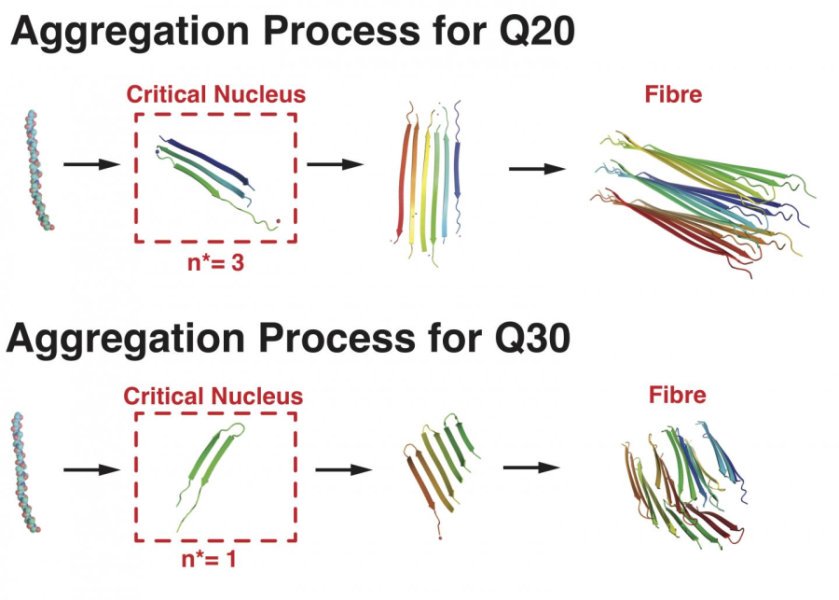
Researchers at Rice University’s Center for Theoretical Biological Physics uncovered an important mechanism in Huntington's disease by creating a computational tool to simulate the behavior of the Huntington's protein, a lynchpin for the manifestation of the disease. Huntington’s disease, a neurodegenerative disease, occurs when the genetic sequence of the Huntington’s protein is passed incorrectly through generations and creates toxic forms of the protein (aggregates). Previous studies have shown that the length of the abnormal sequence is related to the severity of Huntington's disease. The computational tool predicted at what sequence length the toxic aggregates would form, and was able to explain the findings in patients. Computational methods can thus provide novel human-based mechanistic insights into brain-protein related diseases like Alzheimer’s disease and spinocerebellar ataxia.
References
- Chen M, Tsai M, Zheng W, Wolynes PG. The aggregation free energy landscapes of polyglutamine repeats. J Am Chem Soc. 2016;138:15197-15203. doi: 10.1021/jacs.6b08665








