Crystal Structure Offers Insights into Drug Design for a Cancer Drug Target
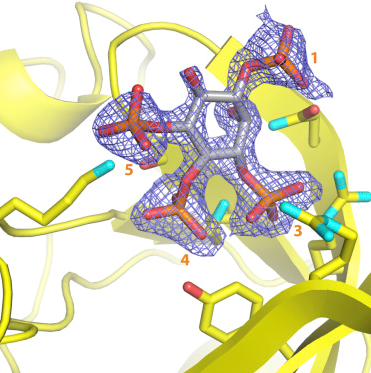
Previously, scientists have identified the protein P-Rex1 as a key driver for cancer progression and metastasis in breast, prostate, and skin cancers. It was found to be produced at high levels in these cancers. Its activation by two key molecules allows cancer cells to spread to other sites. Although it is an attractive drug target, designing molecules to inhibit its activation is challenging without knowing its molecular structure. Researchers at the University of Michigan used a technique called X-ray crystallography to determine the 3-D molecular structure of this enzyme bound to one of its key activating molecules as well as to some of its binding partners. These structures not only provide key insights into how this enzyme is regulated but also reveal key molecular interactions required for the activity of P-Rex1 to guide future small molecule drug design to inhibit cancer metastasis.
References
- Cash JN, Davis EM, Tesmer JJ. Structural and biochemical characterization of the catalytic core of the metastatic factor P-Rex1 and its regulation by PtdIns(3,4,5)P3. Structure. 2016. DOI: 10.1016/j.str.2016.02.022. http://www.cell.com/structure/fulltext/S0969-2126%2816%2900096-4. Accessed April 20, 2016.








